By Megan Bean
Dr O’Donnell’s Advanced Topics course on Regulated Protein Degradation
The Hsp70 system is an essential component of chaperone activity in many organisms. Hsp70 functions include: protein folding, aggregation prevention, trafficking, and enzyme regulation. Hsp70’s ability to bind such a vast array of substrates suggests wide range of conformational plasticity. By utilizing a single mode optical tweezers technique, Mashaghi1 et al., confirms previous theories Hsp70 binds and stabilizes extended peptide segments but also partially folded and near-native protein.
Hsp70 is a highly conserved essential chaperone protein involved in many key cellular events including: folding/unfolding of proteins, assembly of subunits of protein complexes, sorting to subcellular compartments, signaling, protection against stress/apoptosis, recognition of aberrant/unfolded proteins, prevention of aggregation and cell-cycle control (1,4-6,9). Hsp70 interacts with nascent, misfolded, partially folded, almost native proteins and also aggregated proteins (8). Although it is known what domains of Hsp70 are important for binding to these substrates; the actual mechanism and dynamics of how Hsp70 is binding to, recognizing and changing its clients conformation remains elusive (9). The transient nature of the interactions between Hsp70 and its clients, along with the multiple folding stages the clients can be in when binding Hsp70 make assessing these associations challenging. Compounding this difficulty is the addition of co-chaperones and dynamic changes in Hsp70’s ATP hydrolysis cycle. To help surmount these difficulties, Mashaghil et al. employed a single mode optical tweezer technique help generate a much clearer picture of the mechanisms and dynamics of Hsp70-client interactions.
Since its discovery in 1962 Dr. Ferruccio Ritossa Hsp70 has been the focus of extensive study (13). Hsp70’s structure is well-characterized and it has three major domains: an N-terminal ATPase domain responsible for ATP binding, substrate binding domain which forms a groove and is thought to recognize peptide segments that protrude from the various target proteins and finally a C-terminal “lid” domain which can close down on the substrate binding domain in an ATP dependent fashion (14,15).
Our current model of Hsp70 function, based on the work of Mayer and Bukau purposes once entering the ER lumen a newly synthesized peptide segment interacts with Hsp70’s substrate binding domain groove. Once bound, ATP hydrolysis occurs and the C-terminal lid domain clamps down on the groove and peptide segment (Figure 1). It’s important to note that when Hsp70 is alone ATP hydrolysis appears to be slow, however, in the presence of a cochaperone (such as Hsp40) the ATP hydrolysis rate significantly increases (1). This binding of nascent and partially folded proteins is thought to prevent protein aggregation and promote proper folding (1,9,16). The closed Hsp70/ADP-substrate complex is then recognized by a nucleotide exchange factor (GrpE) (1,18-19), and the exchange of ADP for ATP once again opens the lid allowing for the release of the target protein (16). Alternatively Hsp70 also helps remove aberrant proteins by binding to aberrant proteins then passing them onto an E3 ligase, CHIP (Carboxyl-terminus of Hsp70 interacting Protein) for degradation (17). These are a couple ways we know that Hsp70 is interacting with proteins. The question that remains is how Hsp70
is able to bind to so many varying protein conformations?
In a recent issue of Nature a letter by Mashaghi et al.(1) aimed to look at the dynamics of the interactions between Hsp70 and its many client proteins using a single mode optical tweezers technique. Single mode optical fibers “optical tweezers” was developed in Bell Labs by Arthur Ashkin (22). By using a highly focused laser beam to provide an attractive or repulsive force the beam creates an optical trap for objects. For these experiments four maltose binding proteins were linked head to tail (4MBP) to create a fiber that was secured to beads on either end and the beads were suspended in two optical traps(1,2). The bead in one optical trap could be moved closer to the other bead, thereby relaxing the fiber and allowing the MBP proteins to fold. The fibers resistance to being stretched is measured in piconewtons (pN). In the relaxed state the 4MBP were able to fold to their native state. By preforming this in the presence or absence of DnaK (bacterial Hsp70 homolog) the dynamics of the interaction between the folding protein and DnaK could be observed (1). Relaxing, extending and then relaxing 4MBP in the absence of chaperones created a tightly bound protein aggregate. In the presence of the DnaK chaperone system (including DnaJ, GrpE), and ATP nearly 55% of the MBP attained native folding conformation, with tight misfolded protein and some weak misfolded protein (1). This confirmed previous research that suggested that the Hsp70 system could correct refolding of misfolded or aggregated proteins (1,9). In additional optical tweezer experiments MBP was stretched and then relaxed and incubated with DnaK for 5 seconds and stretched again. Surprisingly, MBP incubated with the DnaK would not unfolded suggesting that DnaK was able to recognize the partially folded structure and stabilize it (1). Mashaghi et al. also tested a mutant forms of DnaK with either a truncated lid or lacking the substrate binding groove and found that the lid plays a much larger role in binding than previously thought, a finding that is supported by other current research (12). The lid was essential for recognition of folded/partially folded structures while the groove was essential for binding of non-folded segments (1). When DnaK was in the ATP bound and open conformation the lid and groove appeared to work synergistically to stabilize binding of the substrate. This newly discovered ability to associate with folded/partially folded structures leads to new models for how Hsp70 is recognizing client proteins. For example, Hsp70 may not only be protecting nascent unfolded proteins from aggregation but may also be stabilizing partially folded proteins until they are closer to their native state and then handing them off in later stage chaperones such as Hsp90 and GroEL (11,14). Previous studies demonstrated that Hsp70 could destabilize misfolded proteins and aggregations (21). This may be in part due to its ability to bind to partially folded proteins and prevent or diminish the impending misfolding event. However, it could also be that DnaK is somehow discriminating between a native versus non-native protein. This is something that should be explored along with a reassessment of the role of the co-chaperones (8). This study confirms that addition of the co-chaperone improved in binding efficiency and aggregation prevention yet the role of the co-chaperone was not fully explored (1,12). This leaves the possibility that it is the co-chaperones that allow Hsp70 to distinguish between a native or nonnative structure (Figure 1).
This study showed that in Hsp70 the lid domain and substrate binding groove work synergistically to help Hsp70 interact with its target proteins (1). These studies using a single mode optical tweezers expand our understanding of the dynamics of Hsp70 binding to its substrate. These data further support a model where Hsp70 prevents aggregation and misfolding by binding to partially folded and nascent peptide segments, however they did not show definitively how Hsp70 is working with misfolded proteins. Since misfolded proteins play a critical role in many neurodegenerative diseases the question of how Hsp70 is targeting these proteins is a pertinent one. This study without a doubt demonstrated the power of optical tweezers as a technique to further dissect protein-protein interaction dynamics for chaperones with higher resolution.
Figure 1
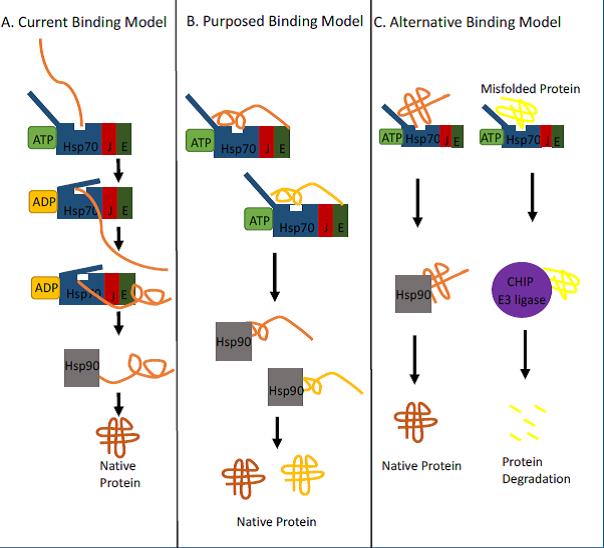
Figure 1. Diverse models for Hsp70 binding to client proteins.
A.) The current, and well supported, model for Hsp70 system binding commences with Hsp70 in an ATP-driven open lid conformation binding to a newly translated protein, where the hydrophobic peptide sequence of the nascent protein interacts with the substrate binding groove. Once bound to Hsp70, ATP hydrolysis changes the conformation to closed and the lid clamps down on to the peptide segment. This model ends with the subsequent transfer of the protein to another chaperone such as Hsp90. It was concluded that this initial binding to the nascent protein prevented protein aggregation. B.) New evidence from Mashaghi et al. suggests another binding possibility for Hsp70 wherein Hsp70 can also recognize partially folded proteins via an open lid conformation. Binding of the lid and substrate binding groove is needed for partially folded proteins. This binding would allow Hsp70 to hold on to the protein longer before passing it off to later stage chaperones and prevent aggregation. C. However, this new evidence could also support a model where Hsp70 system is able to distinguish misfolded proteins from native proteins. Hsp70 could bind to near native proteins helping to stabilize it then passing the protein onto later stage chaperones. If the protein is misfolded, however, it could pass it off to an E3 ligase like CHIP for later degradation.
References
1.) Alireza Mashaghi1, Sergey Bezrukavnikov1, David P. Minde1, Anne S. Wentink, Roman Kityk, Beate Zachmann-Brand,Matthias P. Mayer, Günter Kramer, Bernd Bukau & Sander J. Tans, Alternative modes of client binding enable functional plasticity of Hsp70. Nature. 100, 1-4, (2016).
2.) Philip H. Jones, Onofrio M. Maragò & Giovanni Volpe Optical Tweezers: Principles and Applications
3.) Li Z1, Srivastava P. Heat-shock proteins. Curr Protoc Immunol. 2004 Feb;Appendix 1:Appendix 1T. doi:10.1002/0471142735.ima01ts58. Review.
4.) Normington K, et al. (1989) S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell 57(7):1223-36 PMID: 2661019
5.) Rose MD, et al. (1989) KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57(7):1211-21
6.) Nishikawa SI, et al. (2001) Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol 153(5):1061-70 PMID: 11381090
7.) Matlack KE, et al. (1999) BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell 97(5):553-64
8.) Mayer, M. P. & Bukau, B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684 (2005).
9.) Szabo, A. et al. The ATP hydrolysis-dependent reaction cycle of the Escherichia
coli Hsp70 system DnaK, DnaJ, and GrpE. Proc. Natl Acad. Sci. USA 91,10345–10349 (1994).
10.) Qi, R. et al. Allosteric opening of the polypeptide-binding site when an Hsp70
binds ATP. Nat. Struct. Mol. Biol. 20, 900–907 (2013).
11.) Kityk, R., Kopp, J., Sinning, I. & Mayer, M. P. Structure and dynamics of the
ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874
12.) Marcinowski, M. et al. Substrate discrimination of the chaperone BiP by autonomous and cochaperoneregulated conformational transitions.Nat. Struct. Mol. Biol. 18, 150–158 (2011).
13.) Capocci M, Santoro MG, Hightower LE. The life and times of Ferruccio Ritossa.2014 Cell Stress Chaperones. Sep;19(5):599-604. doi: 10.1007/s12192-014-0525-4.
14.) Zhu, X. et al. Structural analysis of substrate binding by the molecular
chaperone DnaK. Science 272, 1606–1614 (1996).
15.) Rüdiger, S., Germeroth, L., Schneider-Mergener, J. & Bukau, B. Substrate specificity of the DnaK chaperone determined by screening cellulose-boundpeptide libraries. EMBO J. 16, 1501–1507 (1997).
16.) Mayer M P (2010). “Gymnastics of Molecular Chaperones”. Cell. 39: 321–331.
17.) Lüders J, Demand J, Höhfeld J (February 2000). “The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome”. J. Biol. Chem. 275 (7): 4613–7.
18.) Schlecht, R., Erbse, A. H., Bukau, B. & Mayer, M. P. Mechanics of Hsp70
chaperones enables differential interaction with client proteins. Nat. Struct. Mol.
Biol. 18, 345–351 (2011).
19.) Bechtluft, P. et al. Direct observation of chaperone-induced changes in a protein folding pathway. Science 318, 1458–1461 (2007).
20.) Ishiai, M., Wada, C., Kawasaki, Y. & Yura, T. Replication initiator protein RepE of
mini-F plasmid: functional differentiation between monomers (initiator) and dimers(autogenous repressor). Proc. Natl Acad. Sci. USA 91, 3839–3843 (1994).
21.) Sharma, S. K., De los Rios, P., Christen, P., Lustig, A. & Goloubinoff, P. The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nat. Chem. Biol. 6, 914–920 (2010).
22.) Koch MD1, Shaevitz JW2. Introduction to Optical Tweezers. Methods Mol Biol. 2017;1486:3-24.
Cite this article:
Bean, Megan. “Hsp70 Conformational Plasticity Allows for Expansive Chaperone Role.” The D.U.Quark, 2, 1 (2017): 29-34. https://duquark.com/2017/06/26/hsp70-conformational-plasticity-allows-for-expansive-chaperone-role/
Download the PDF here




Leave a comment